Genetic Algorithm for 2D Multi-Element Optimization
Formula SAE2D CFDAerodynamicsSoftware DevelopmentThe goal of this project was to improve upon the brute-force method used on EV5 for finding the optimal multi-element airfoil configurations.
Due to this being a multi-variable optimization problem where the number of variables is n = 2 + 4a where a is the number of airfoils, a genetic algorithm is the appropriate tool to use here as the number of variables is more than 3.
The following serves as a breakdown of the program structure. A brief performance comparison between the brute-force and genetic optimization methodologies is also included at the end.
Program Structure
For the purpose of airfoil optimization, we want to determine the optimal scale, angle of attack, x-position, and y-position of each airfoil relative to one another. It has already been determined in Justification for Aerodynamics that lap time is more sensitive to downforce than drag, therefore the metric used to define fitness will be the lift coefficient of the given multi-element setup. The goal of the algorithm will be to find the highest lift coefficient possible within the defined number of generations. A high-level overview of the algorithm can be seen below in Figure 1.
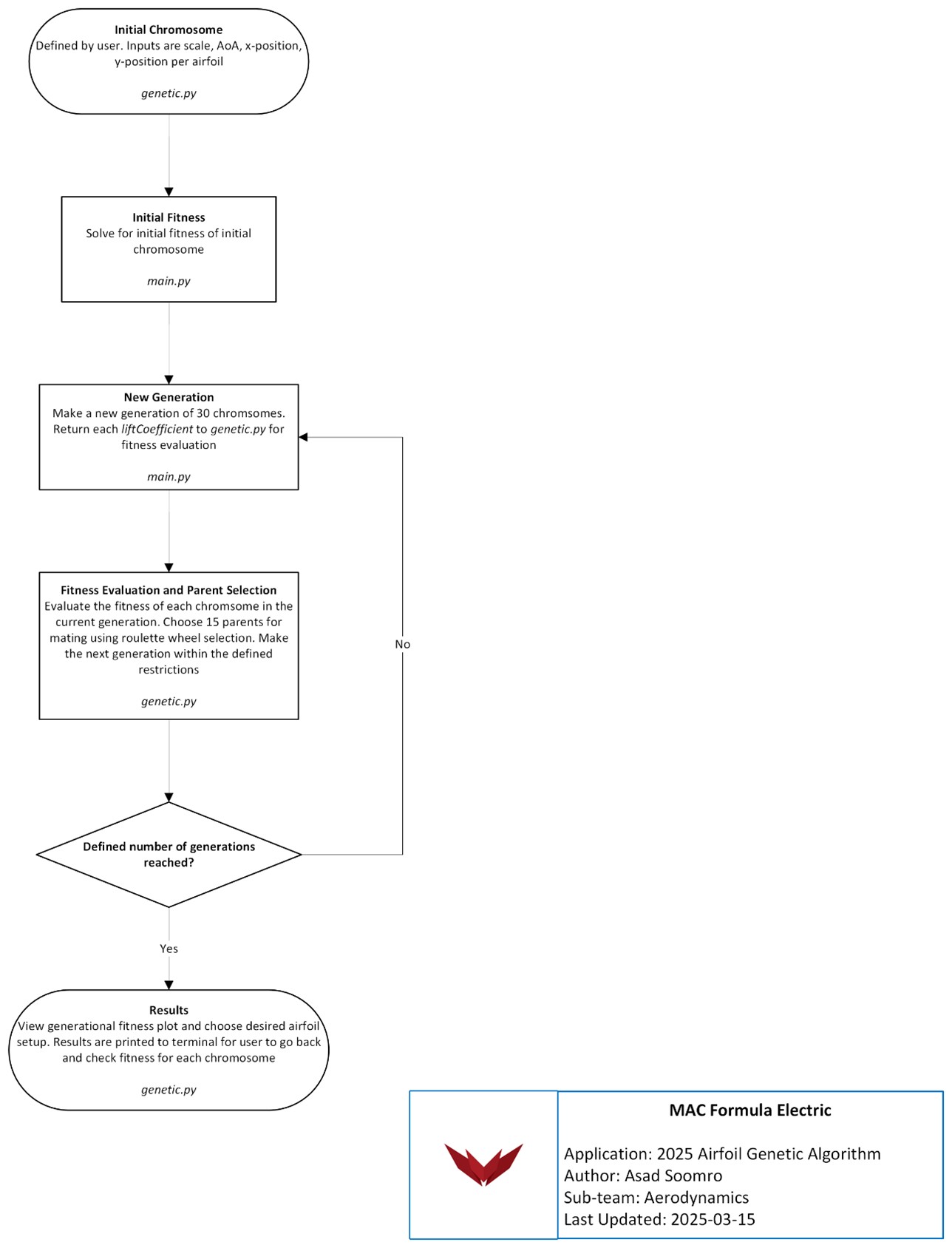
Fig. 1 2025 airfoil genetic algorithm
The genetic algorithm is split into 2 files, main.py and genetic.py. The CFD calculations are handled by main.py through MSES, while the fitness evaluation, parent selection, and generation creation are handled by genetic.py through pygad.
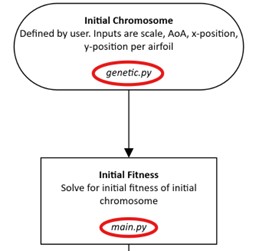
Fig. 2 genetic.py and main.py
The first thing to note is pygad was selected due to its easy implementation of custom fitness functions, and due to it also being written in python which is one of the most commonly known and used programming languages. Other GA libraries can be used if desired, however I felt pygad would be easily scalable for the reasons described.
The program takes in an initial chromosome to prevent startup trauma on both the CFD and GA sides of the program. A liftCoefficient variable is initialized to store the current lift coefficient.
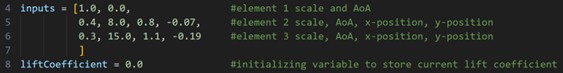
Fig. 3 Initial inputs to GA
An instance of pygad is then created, and the algorithm starts running by solving for the initial inputs described above.
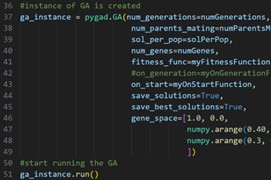
Fig. 4 Instance of GA is created and run
The first thing to run is the on_start function. This is an optional function within pygad that can be used to run after ga_instance.run() is called. For our case, we are using it to evaluate the lift coefficient of the initial set of inputs.
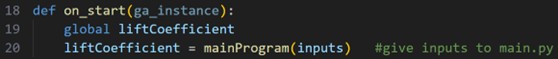
Fig. 5 Inputs are given to main.py for CFD analysis
The inputs are fed into the function mainProgram(inputs) in main.py. This is the function which takes the inputs, runs the MSES CFD, then outputs the result.

Fig. 6 Function to take inputs. Airfoils are coordinate files
The inputs are then parsed into their respective variables to be used by the MSES programs. They are printed to the terminal for user convenience.

Fig. 7 Parsed inputs. Remember python is a 0-index language, which is why the first element of the array is referenced as newInputs[0]
From here, the MSES collection of programs begins to process the inputs and prepare them for CFD analysis. The specific programs used, the documentation for what each of them does, and the relevant code snippets can be found in the tables below.
| Program | Documentation |
| airset.exe | airset.pdf (page 20) |
| mset.exe | mses.pdf (pages 20-24) |
| makespecfile.exe | N/A |
| mses.exe | mses.pdf (pages 24-25) |
| mpolar.exe | mses.pdf (page 27) |
Table 1 MSES programs and their corresponding documentation
| Program | Code Snippet (main.py) |
| airset.exe |  |
| mset.exe |  |
| makespecfile.exe |  |
| mses.exe | 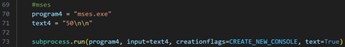 |
| mpolar.exe | 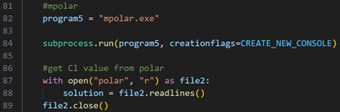 |
Table 2 MSES programs and their corresponding code snippets
At the end of main.py we return liftCoefficient to genetic.py to evaluate the fitness of the current chromosome.

Fig. 8 Return liftCoefficient
Now that the first loop is finished, a new set of inputs are generated from the defined gene_space. The gene space is simply the “bounding box” of physical restrictions that your chromosome can take on. It defines the limits of what the scale, AoA, x-position, and y-position can be for the next generated chromosome.

Fig. 9 Each row of gene_space generates a scale, AoA, x-position, and y-position for the desired number of airfoil elements. Simply increase the number of rows to add more airfoils to the chromosome
The next population of chromosomes are then run through fitnessFunction until the entire population’s fitness has been evaluated. The program ends when the defined number of generations has been reached.
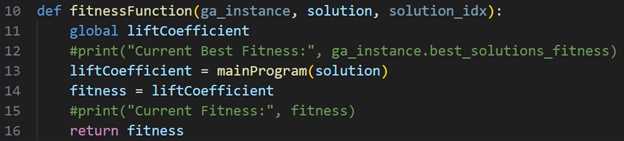
Fig. 10 Evaluate each chromosome’s fitness within the current population. Repeat for the defined number of generations
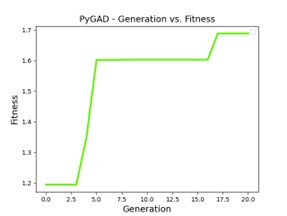
Below is a sample output of the results from the program.
 |
 |
Figs. 11 and 12 The outputs will be a plot of fitness evolution, and as the program is running the user will see the chromosome being evaluated and its fitness (i.e. the lift coefficient)
Brute-Force versus Genetic Optimization
Overall, EV6 benefitted greatly for both the front and rear wing designs. It’s worth noting when moving from EV5 to EV6 that both front and rear wings went from 2 to 3 elements, which also played a great role in improving downforce. However, it is the natural selection of the algorithm which enabled us to ultimately choose the highest downforce configurations in the end.
The 2D CFD results from Ansys Fluent can be seen in the table below. Reference areas are the global chord lengths of the respective multi-element setups.
| Device | EV5 Cl (-) | EV6 Cl (-) | Percentage Increase (%) |
| Front Wing | -1.72 | -2.71 | 57.56 |
| Rear Wing | -1.93 | -3.35 | 73.58 |
Table 3 EV5 versus EV6 front and rear wing lift coefficients
The code and some sample airfoil coordinates are available at this repository under a MIT license. Note that only my code is included in the repository, and not the MSES collection of programs, since those were specially given to MAC Formula Electric through an academic license.
You can find additional references for pygad and all of its functions here.